PINNet: A deep neural network with pathway prior knowledge for Alzheimer’s disease
More Links
Enhancing AI Interpretability with Biological Pathway Knowledge: Introducing PINNet
Machine learning models have revolutionized our ability to analyze complex biological data, but often they operate as “black boxes,” offering little insight into how they reach their conclusions. To address this challenge, we’ve developed a novel deep neural network called PINNet—a model specifically designed to incorporate biological prior knowledge directly into its architecture. PINNet’s unique approach not only enhances predictive accuracy but significantly improves interpretability, giving clear insights into the underlying biological mechanisms involved in diseases.
Why Prior Knowledge Matters
Traditional neural networks treat every feature (e.g., gene expression levels) equally, ignoring established biological relationships. In contrast, PINNet leverages well-curated biological pathway databases—such as Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG)—to guide its learning process. This incorporation of biological prior knowledge helps PINNet pinpoint precisely which genes and pathways are most influential in its predictions.
How PINNet Integrates Pathway Knowledge
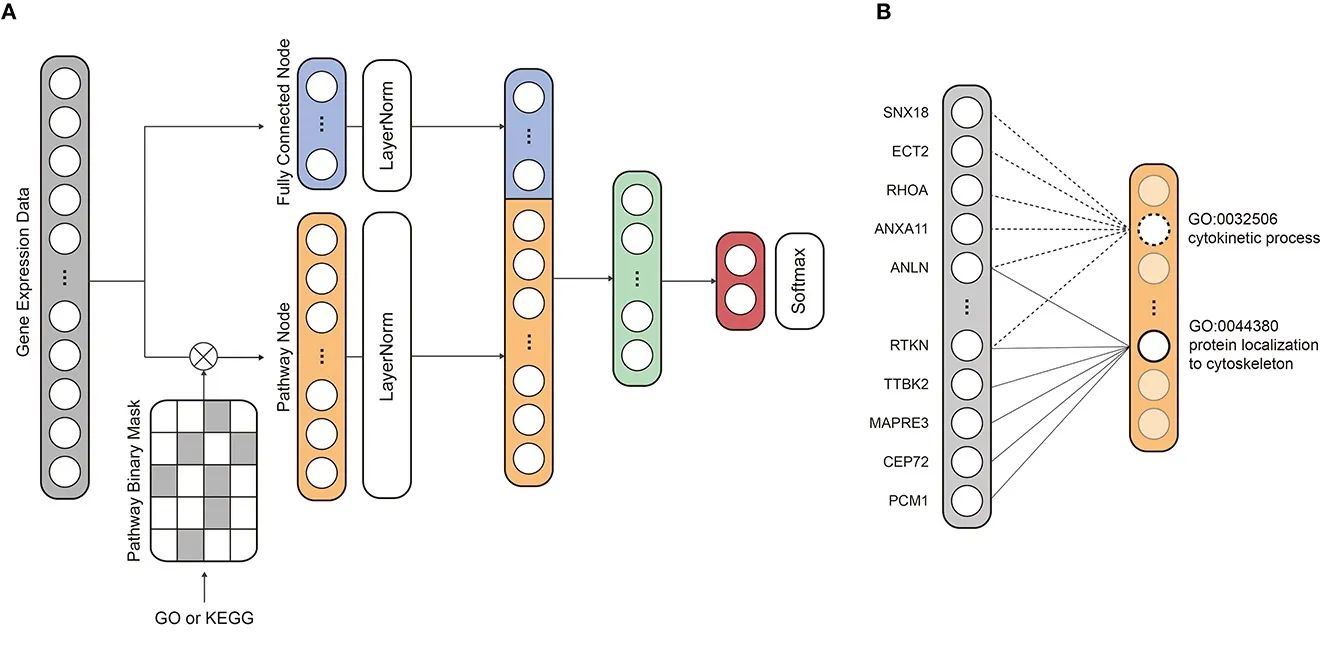
PINNet integrates biological pathway information directly into its neural network structure. It uses a special pathway layer that links individual genes to known biological pathways, ensuring that the neural network learns not just from raw data but from biologically meaningful relationships.
Specifically, PINNet employs:
- A pathway layer, where connections reflect known gene-pathway relationships.
- Normalization methods to ensure fairness in evaluating pathway importance, irrespective of the number of genes involved.
- Deep learning interpretation techniques (DeepSHAP) to identify critical genes and pathways influencing the predictions.
Key Insights from PINNet
Applying PINNet to transcriptomic data provided exciting results:
- Enhanced interpretability: PINNet consistently identified biologically significant pathways linked to immune response, neural plasticity, and cell signaling, providing deep insights into disease mechanisms.
- Gene-level insights: PINNet highlighted key genes known to play roles in critical disease processes, confirming its biological relevance and effectiveness.
Broader Impact and Future Directions
PINNet’s innovative use of biological prior knowledge sets a new benchmark for interpretability in AI-driven biological research. Beyond disease prediction, this methodology can broadly impact how we use AI to understand complex biological systems across numerous conditions.
We believe that integrating known biological structures directly into machine learning models is a transformative step, paving the way for more informed, precise, and actionable scientific discoveries.
For detailed methodology and findings, read our full paper here.